Update March 2020: You can find a very interesting comparison between data.tableand dplyr here.
Introduction
I have recently noticed that every R script I wrote starts with library(data.table). And that seems a very compelling reason for me to write a post about it.
You may have seen that as the machine learning community is developing, Python has gained enormous popularity and as a result, the Pandas library has automatically become a staple for a lot of people.
However, if I had a choice, I would definitely prefer using R data.table for relatively large dataset data manipulation, in-depth exploration and ad-hoc analyses. Why? Because it’s so fast, elegant and beautiful (sorry if I got too enthusiastic!). But hey, working with data means you’ll have to turn the code upside down, mould it, clean it for maybe… uhm… a thousand times before you can actually build a machine learning model. In fact, data pre-processing usually takes 80–90% the time of a data science project. That’s why I can’t afford slow and inefficient code (and I don’t think anyone should, as a data professional).
So let’s dive into what data.table is and why many people have become big fans of it.
1. So what the heck is data.table?
data.table package is an extension of data.frame package in R. It is widely used for fast aggregation of large datasets, low latency add/update/remove of columns, quicker ordered joins, and a fast file reader.
That sounds good, right? You may think it’s difficult to pick up, but actually a data.table is also a type of data.frame. So rest assured that any code you use for a data.frame will also work as well with a data.table. But believe me, once you’ve used data.table, you’ll never want to use the base R data.frame syntax again. Keep reading if you want to know why.
2. Data.table is extremely fast
From my own experience, working with fast code can really improve the thinking flow in the data analysis process. Speed also very important in a data science project, in which you usually have to quickly prototype an idea.
When it comes to speed, data.table puts all other packages in Python and many other languages to shame. This is shown in this benchmark, which compares tools from R, Python and Julia. To do five data manipulations on a 50GB dataset, data.table only took on average 123s, while Spark took 381s, (py)datatable took 712s, and pandas could not do the task due to out of memory.
One of the most powerful functions in data.table package is fread(), which imports data similarly to whatread.csv() does. But it’s optimized and much much faster. Let’s look at this example:require("microbenchmark")
res <- microbenchmark(
read.csv = read.csv(url("https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data"), header=FALSE),
fread = data.table::fread("https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data", header=FALSE),
times = 10)
res
The results will show that on average, fread() will be 3-4 times faster than read.csv() function.
3. It is intuitive and elegant
Almost every data manipulation with data.table will look like this:
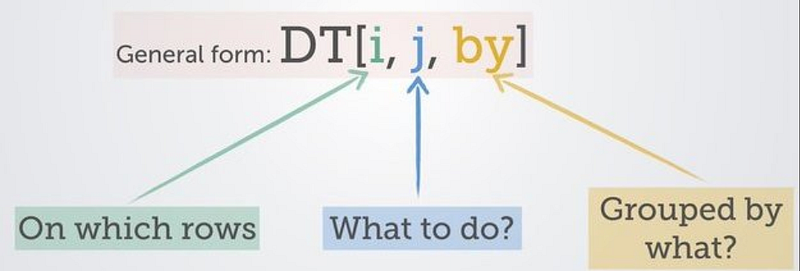
As a result, the code you write will be very consistent and easy to read. Let’s take the US Census Income dataset for illustration purposes:dt <- fread("https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data", header=FALSE)
names(dt) <- c("age", "workclass", "fnlwgt", "education", "education_num", "marital_status", "occupation", "relationship", "race", "sex", "capital_gain", "capital_loss","hours_per_week", "native_country", "label")
In the below examples, I’ll compare the code in base R, Python and data.table, so that you can easily compare them:
- To compute the average
ageof all “Tech-support” workers:
> in base R you’d probably write something like this:mean(dt[dt$occupation == 'Tech-support', 'age'])
> in Python:np.mean(dt[dt.occupation == 'Tech-support'].age)
> vs. in data.table:dt[occupation == 'Tech-support', mean(age)]
As you can see in this simple example, data.table removes all redundancy of repeating dt all the times, compared to Python and base R. This in turn reduces the chance of making typo mistakes (remember the coding principle DRY — Don’t repeat yourself!)
2. To aggregate age by occupation for all male workers:
> in base R you’d probably write:aggregate(age ~ occupation, dt[dt$sex == 'Male', ], sum)
> in Python:dt[dt.sex == 'Male'].groupby('occupation')['age'].sum()
> vs. in data.table:dt[sex == 'Male', .(sum(age)), by = occupation]
The by = term defines which column(s) you want to aggregate your data on. This data.table syntax may seem a little intimidating at first, but once you get used to it you’ll never bother typing “groupby(…)” again.
3. To conditionally modify values in a column, for example to increase the age of all Tech-support workers by 5 (I know it’s a silly example, but just for illustration :)).
> in base R you’d probably have to write this horrible line (who has time to write dt$ so many times!):
dt$age[dt$occupation ==> in Python (there are several alternatives that are equally long):
mask = dt['occupation'] ==or using np.where:
dt['occupation'] ==> vs. in data.table:dt[occupation == 'Tech-support', age := age + 5]# and the ifelse function does just as nicely:
dt[, age := ifelse(occupation == 'Tech-support', age + 5, age)]
It’s almost like a magic, isn’t it? No more cumbersome repetitive code, and it keeps yourself DRY. You may have noticed the strange operator := in the data.table syntax. This operator is used to assign new values to an existing column, just like using the argument inplace=True in Python.
4. Renaming columns is a breeze in data.table. If I want to rename occupation column as job:
> in base R you might write:
colnames(dt)[which(names(dt) == "occupation")] <- "job"> and in Python:
dt = dt.rename(columns={'occupation': 'job'})> vs. in data.table:setnames(dt, 'occupation', 'job')
5. What about applying a function to several columns? Suppose you want to multiply capital_gain and capital_loss by 1000:
> in base R:apply(dt[,c('capital_gain', 'capital_loss')], 2, function(x) x*1000)
> in Python:dt[['capital_gain', 'capital_loss']].apply(lambda x : x*1000)
> vs. in data.table:dt[, lapply(.SD, function(x) x*1000), .SDcols = c("capital_gain", "capital_loss")]
You might find this syntax in data.table is a little quirky, but the .SDcols argument comes in very handy in many cases, and the general form of data.table syntax is preserved this way.
Conclusion:
From a few simple illustrations above, one can see that the code in R data.table is in many cases faster, cleaner and more efficient than in base R and Python. The form of data.table code is very consistent. You only need to remember:
DT[i, j, by]As an additional note, the data subsetting with i by keying a data.table even allows faster subsets, joins and sorts, which you can read more about in this documentation, or this very useful cheat sheet.
Thank you for reading. If you like this post, I would write more about how to do advanced data wrangling with R and Python in the future posts.

